Logistic Regression
Session 10
(small modifs by Kim Antunez & ChatGPT)
2024-04-16
Non-linearity
Feedback about Exam 1
- Structure of code nearly perfect :
install.packagesin comments.setwdOUTSIDE thedatafolder
select,group_by,mutatemastered in all groups- Don’t forget to explain each step (
paste0,as.Dateorwdaysometimes forgotten) - Q8 and Q9 not right for everybody
➡️ All marks are > 15/20. BRAVO! But next exams will be more challenging 😳
Non linearity for variables
Variables that violate model assumptions will often do so due to high skewness (non symetric distribution)
A possible fix might be to transform the scale on which those variables are measured.
Often:
- logarithmic transformation (predictor will have a quadratic effect on the dependent variable)
- square transformation (more complex to interpret)
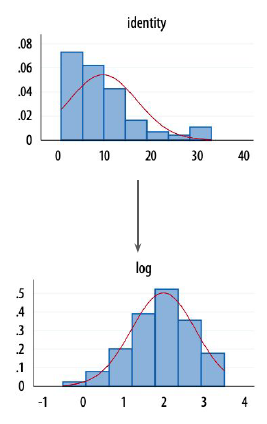
Logarithmic coefficients
linear relationship (last week) Y=α+βX
log-linear relationship lnY=α+βX
lin-log relationship Y=α+βlnX
log-log relationship lnY=α+βlnX
An increase in 1 unit of X is associated with an increase of β unit in Y
An increase in 1 unit of X is associated with a 100 β% change in Y
A 1% increase in X is associated with a 0.01 β% change in Y
A 1% increase in X is associated with a β% change in Y
Generalized Linear Models (GLM)
Nonlinearity for models
When dealing with a binary dependent variable (Y=0 or Y=1), logistic regression is commonly used.
In logistic regression, we introduce a link function to model the relationship between the predictor(s) and the binary response variable.
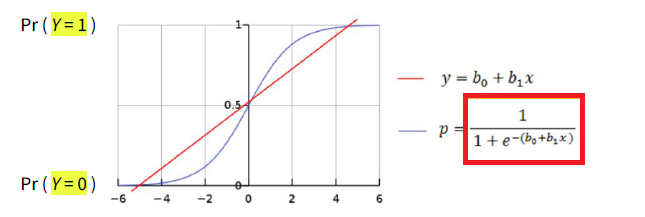
It allows to express the relationship between Y and its predictor(s) through a linear model, at the price of having to interpret that relationship through log-odds, which can be converted into probabilities of P(Y=1) through exponentiation (see after).
Logistic regression coefficients
Logistic regression equation
L=ln(p1−p)⏟= log-odds = logit =α+βX or p=eα+βX1+eα+βX
with
- p=P(Y=1)
- ln : natural logarithm.
- X is the predictor variable.
- α is the intercept.
- β is the coefficient associated with the predictor variable.
Interpretation : odd-ratios
Odds Ratio=eβ
For 1-unit increase in X, P(Y=1) increase by a factor of eβ.
- OR>1 denotes a higher probability of Y = 1 (OR = 1.24 denotes a 24% increase)
- OR<1 denotes a lower probability of Y = 1 (OR = 0.5 denotes P(Y=1) divided by 2)
Before Exercise 1
Testing for the presence of the bacterium H. influenzae (y) in children based on the administration of a medication versus a placebo (trt). The presence of the virus was monitored at weeks 0, 2, 4, 6, and 11 (week).
'data.frame': 220 obs. of 6 variables:
$ y : Factor w/ 2 levels "n","y": 2 2 2 2 2 2 1 2 2 2 ...
$ ap : Factor w/ 2 levels "a","p": 2 2 2 2 1 1 1 1 1 1 ...
$ hilo: Factor w/ 2 levels "hi","lo": 1 1 1 1 1 1 1 1 2 2 ...
$ week: int 0 2 4 11 0 2 6 11 0 2 ...
$ ID : Factor w/ 50 levels "X01","X02","X03",..: 1 1 1 1 2 2 2 2 3 3 ...
$ trt : Factor w/ 3 levels "placebo","drug",..: 1 1 1 1 3 3 3 3 2 2 ...In mean, bacteria presence decreases with time
0 1
0 0.10000000 0.90000000
2 0.09090909 0.90909091
4 0.26190476 0.73809524
6 0.27500000 0.72500000
11 0.27272727 0.72727273In mean, bacteria presence decreases thanks to drugs
Fit models (most parsimonious to full).
Note: If you forget to specify family = binomial in the glm function for a binary response variable (i.e., a binary logistic regression), the default family assumed is family = gaussian, which corresponds to linear regression.
Compare them.
=====================================================
Model 1 Model 2 Model 3
-----------------------------------------------------
(Intercept) 1.41 *** 1.96 *** 2.55 ***
(0.17) (0.29) (0.41)
week -0.11 * -0.12 **
(0.04) (0.04)
trtdrug -1.11 **
(0.43)
trtdrug+ -0.65
(0.45)
-----------------------------------------------------
AIC 219.38 214.91 211.81
BIC 222.77 221.70 225.38
Log Likelihood -108.69 -105.45 -101.90
Deviance 217.38 210.91 203.81
Num. obs. 220 220 220
=====================================================
*** p < 0.001; ** p < 0.01; * p < 0.05GLM2 (Model 3) minimizes AIC and seems to be the best model.
Exponentiate GLM2 coefficients to read log-odds.
# A tibble: 4 × 5
term estimate std.error statistic p.value
<chr> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 12.8 0.406 6.28 3.42e-10
2 trtdrug 0.331 0.425 -2.60 9.25e- 3
3 trtdrug+ 0.521 0.446 -1.46 1.44e- 1
4 week 0.891 0.0441 -2.62 8.72e- 3For 1-unit increase in
week, P(Y=1) is divided by1/0.89 = 1.12.Taking resp. drug/drug+ instead of a placebo, P(Y=1) is divided by 3 and 2.
Exam 2
- Instructions here
- in group & graded (like Exam 1)
- same real-world dataset (road traffic accidents in France)
- data visualisation and data analysis (correlation, link between variables) tasks
- Submit a script called
exam2_solution.R(Subject : “DSR Exam 2 Submission”, To : kim.antunez@sciencespo.fr) - Deadline : apr. 30th, before 7:15 pm.
- Make sure your script is well-organized (you all did well last time)
Exam 2
- Instructions here
- in group & graded (like Exam 1)
- same real-world dataset (road traffic accidents in France)
- data visualisation and data analysis (correlation, link between variables) tasks
- Submit a script called
exam2_solution.R(Subject : “DSR Exam 2 Submission”, To : kim.antunez@sciencespo.fr) - Deadline : apr. 30th, before 7:15 pm.
- Make sure your script is well-organized (you all did well last time)